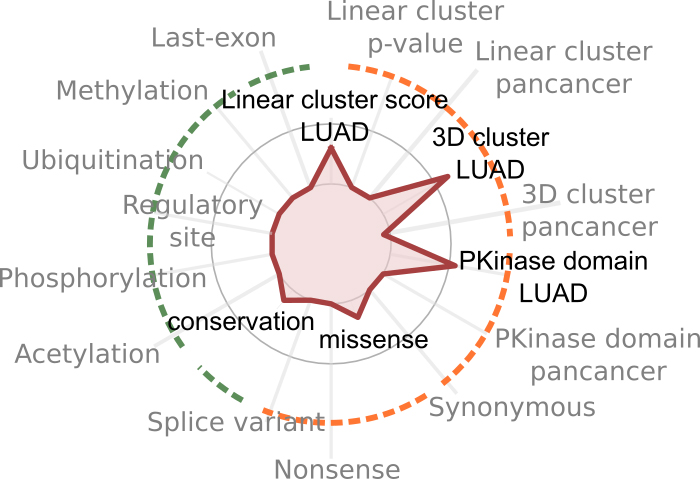
Loading...
-
We are a research group dedicated
to the computational study of cancer
at the genomic levelOur lab is integrated in the Institute for
Research Biomedicine (IRB) in Barcelona -
-
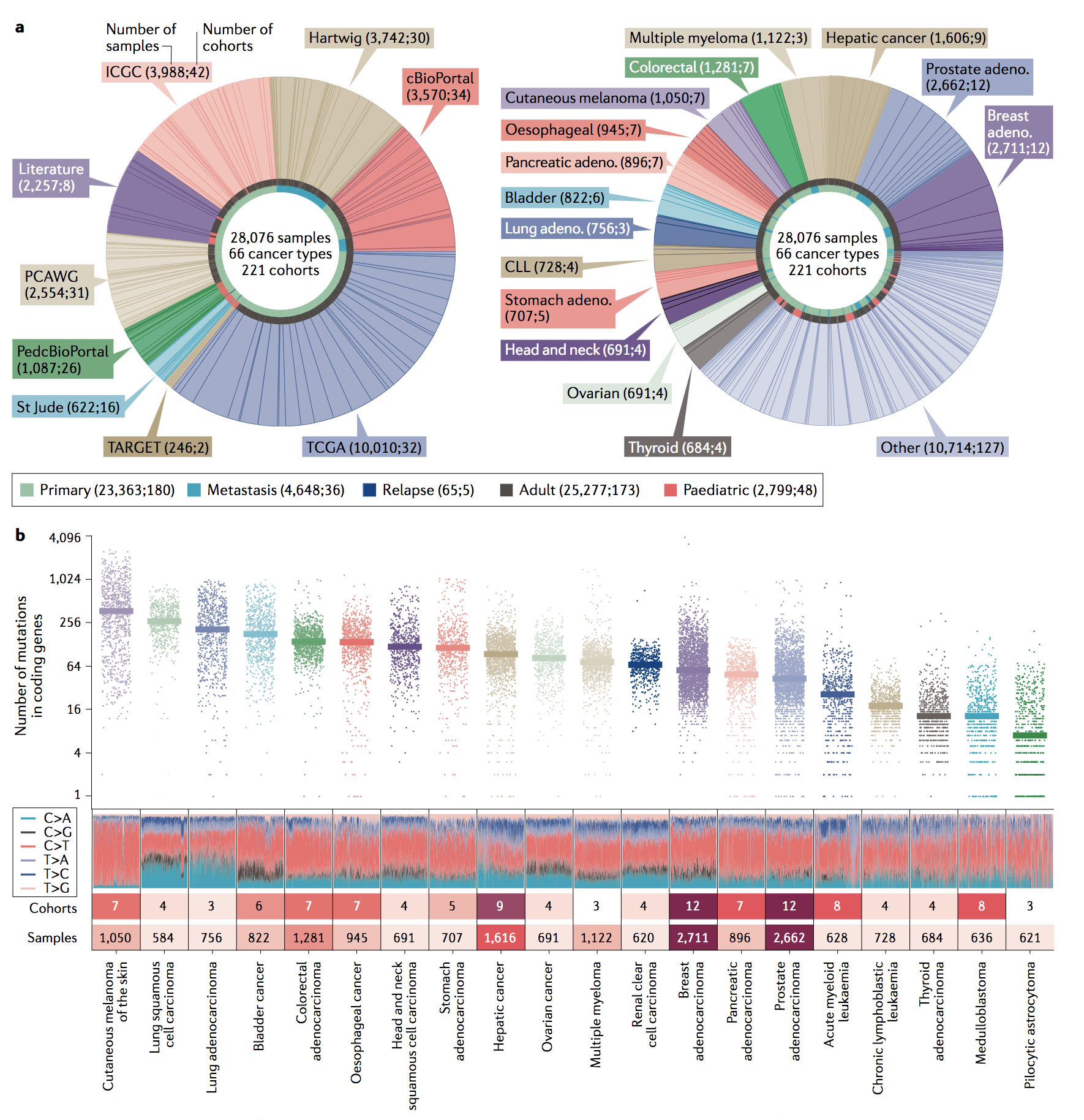
IntOGen release (2023)
We are pleased to announce that a
new release of intOGen is out.

© 2023 BBGLab